| dyads_test_vs_ctrl_m1_shift0 (dyads_test_vs_ctrl_m1) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
; dyads_test_vs_ctrl_m1; m=0 (reference); ncol1=14; shift=0; ncol=14; wwgtGGCCCAAAww
; Alignment reference
a 344 277 193 171 69 58 79 52 62 991 895 813 389 279
c 217 215 102 93 20 16 872 960 953 39 73 95 228 254
g 215 232 572 89 958 972 70 21 32 20 44 76 156 220
t 317 369 226 740 46 47 72 60 46 43 81 109 320 340
|
| LOC_Os02g42380_M1657_1.02_CISBP_shift1 (LOC_Os02g42380:M1657_1.02:CISBP) |
 |
0.706 |
0.504 |
3.479 |
0.896 |
0.892 |
2 |
3 |
4 |
1 |
1 |
2.200 |
1 |
; dyads_test_vs_ctrl_m1 versus LOC_Os02g42380_M1657_1.02_CISBP (LOC_Os02g42380:M1657_1.02:CISBP); m=1/6; ncol2=10; w=10; offset=1; strand=D; shift=1; score= 2.2; -kkGGgcCCmm---
; cor=0.706; Ncor=0.504; logoDP=3.479; NsEucl=0.896; NSW=0.892; rcor=2; rNcor=3; rlogoDP=4; rNsEucl=1; rNSW=1; rank_mean=2.200; match_rank=1
a 0 18 7 0 0.0000 13 11 0.0000 7 45 28 0 0 0
c 0 13 15 0 0.0000 10 66 1.0000 93 33 41 0 0 0
g 0 41 33 93 1.0000 66 10 0.0000 0 15 13 0 0 0
t 0 28 45 7 0.0000 11 13 0.0000 0 7 18 0 0 0
|
| TCP23_TCP23_ArabidopsisPBM_shift2 (TCP23:TCP23:ArabidopsisPBM) |
 |
0.729 |
0.521 |
5.457 |
0.880 |
0.855 |
1 |
2 |
1 |
4 |
4 |
2.400 |
2 |
; dyads_test_vs_ctrl_m1 versus TCP23_TCP23_ArabidopsisPBM (TCP23:TCP23:ArabidopsisPBM); m=2/6; ncol2=10; w=10; offset=2; strand=D; shift=2; score= 2.4; --GGGGCCCACA--
; cor=0.729; Ncor=0.521; logoDP=5.457; NsEucl=0.880; NSW=0.855; rcor=1; rNcor=2; rlogoDP=1; rNsEucl=4; rNSW=4; rank_mean=2.400; match_rank=2
a 0 0 2 0 1 3 1 0 1 96 1 74 0 0
c 0 0 5 0 1 9 97 98 98 1 96 14 0 0
g 0 0 77 97 98 84 1 2 0 3 1 2 0 0
t 0 0 16 3 0 4 1 0 1 0 2 10 0 0
|
| OJ1581_MA1031.1_JASPAR_shift1 (OJ1581:MA1031.1:JASPAR) |
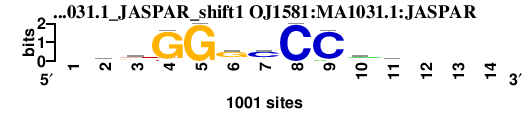 |
0.705 |
0.504 |
3.474 |
0.896 |
0.891 |
3 |
4 |
5 |
2 |
2 |
3.200 |
3 |
; dyads_test_vs_ctrl_m1 versus OJ1581_MA1031.1_JASPAR (OJ1581:MA1031.1:JASPAR); m=3/6; ncol2=10; w=10; offset=1; strand=D; shift=1; score= 3.2; -kkGGgcCCmm---
; cor=0.705; Ncor=0.504; logoDP=3.474; NsEucl=0.896; NSW=0.891; rcor=3; rNcor=4; rlogoDP=5; rNsEucl=2; rNSW=2; rank_mean=3.200; match_rank=3
a 0 175 74 0 0 127 112 0 71 445 279 0 0 0
c 0 131 146 0 0 101 661 1000 929 335 414 0 0 0
g 0 414 335 929 1000 661 101 0 0 146 131 0 0 0
t 0 279 445 71 0 112 127 0 0 74 175 0 0 0
|
| TCP16.ampDAP_M0753_AthalianaCistrome_rc_shift0 (TCP16.ampDAP:M0753:AthalianaCistrome_rc) |
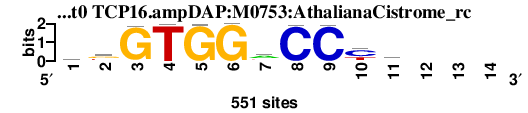 |
0.700 |
0.550 |
2.022 |
0.895 |
0.878 |
6 |
1 |
6 |
3 |
3 |
3.800 |
4 |
; dyads_test_vs_ctrl_m1 versus TCP16.ampDAP_M0753_AthalianaCistrome_rc (TCP16.ampDAP:M0753:AthalianaCistrome_rc); m=4/6; ncol2=11; w=11; offset=0; strand=R; shift=0; score= 3.8; wkGTGGaCCyr---
; cor=0.700; Ncor=0.550; logoDP=2.022; NsEucl=0.895; NSW=0.878; rcor=6; rNcor=1; rlogoDP=6; rNsEucl=3; rNSW=3; rank_mean=3.800; match_rank=4
a 160 89 11 0 2 0 307 3 0 38 240 0 0 0
c 92 24 1 4 2 0 39 546 545 321 69 0 0 0
g 88 242 539 0 547 551 82 2 0 9 161 0 0 0
t 211 196 0 547 0 0 123 0 6 183 81 0 0 0
|




